After a dozen years as a Chaired Professor and Howard Hughes Medical Institute Investigator at The Rockefeller University and a decade working in the biopharmaceutical industry, Stephen K. Burley, M.D., D. Phil., attempted to retire in 2012. “Within months, it became clear that I was failing at retirement,” says Burley. “Fortunately, Rutgers University—specifically Drs. Ken Breslauer and Chris Molloy—came to the rescue and offered me the opportunity to succeed Professor Helen M. Berman as Director of the Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) and establish a new interdisciplinary research center on the Busch Science Campus.”
Dr. Stephen K. Burley is University Professor and Henry Rutgers Chair; Founding Director, Institute for Quantitative Biomedicine, Director (IQB), RCSB Protein Data Bank (PDB), and Co-Lead, Cancer Pharmacology Research Program, Rutgers Cancer Institute of New Jersey, The State University of New Jersey. As an expert in bioinformatics, structural biology, proteomics, clinical medicine/oncology, and structure/fragment based drug discovery, Burley has authored/coauthored more than 340 scholarly scientific articles that explore these disciplines. He has received many honors and awards for his high level of excellence in scientific research, education, and leadership.
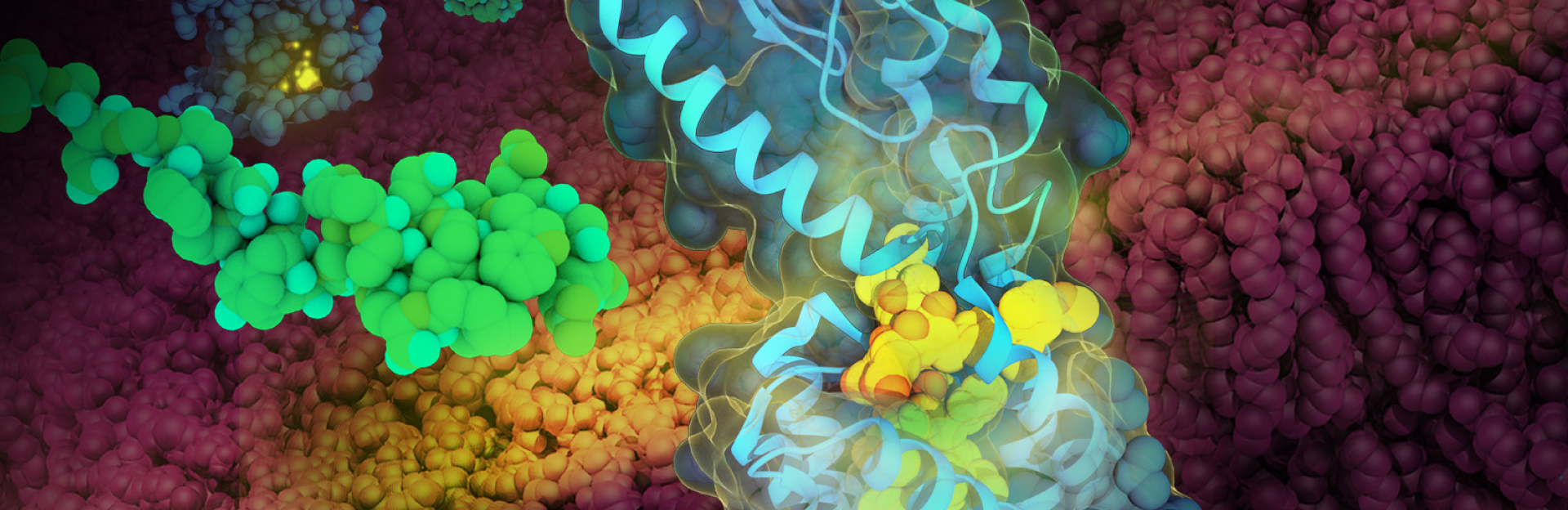
GLP-1 Receptor Recognizing a GLP-1 Analog by Maria Voigt.
Glucagon-Like Peptide-1 (GLP-1) receptor (blue, PDB structure 5NX2) recognizing a GLP-1 analog (yellow), with liraglutide (green, from PDB structure 4APD) free in solution. The cell membrane is shown in red. Open access to these and related structures in the PDB will facilitate discovery and development of new treatments for diabetes.
PDB structures are the molecules of life, coming from every Kingdom. Knowledge of 3D structures (shapes) of biomolecules helps to explain how they function in nature, accelerating discovery across the sciences. PDB data impact basic and applied research on health and disease of humans, animals, and plants; production of food and energy; and other research pertaining to global prosperity, climate change, and environmental sustainability. Global open access to PDB data has broadly and deeply impacted fundamental biology, biomedicine, energy sciences, and biotechnology.
The Evolution of Research & Innovation
As each decade passes and new advanced technologies and compute resources are introduced, research and innovation are propelled forward in valuable and inspiring ways. “Since I became a tenure-track Assistant Professor in late 1990, I have witnessed five transformative changes,” shares Burley. “First, structural biology has become a mainstream subdiscipline of the biosciences, capable of delivering experimentally-determined three-dimensional (3D) structure information that explains how biomolecules work. Second, the open-access Protein Data Bank (PDB) has become the single global repository for 3D biostructure information. Third, computers are now much more powerful; helping us store and analyze data more efficiently, solve problems and overcome societal challenges, and spur international cooperation through shared knowledge. Fourth, the research community has embraced open access to data and the FAIR Principles of Findability, Accessibility, Interoperability, and Reusability. The fifth important change is the application of Artificial Intelligence/Machine Learning (AI/ML) methods to predicting 3D structures of proteins, solely from knowledge of amino acid sequence with accuracies comparable to low resolution experimental methods.”
For those interested in starting a research career in structural biology, Dr. Burley recommends pursuing cryo-electron tomography (cryo-ET). “This rapidly evolving method bridges the gap between light microscopy and in vitro structure determination methods, enabling atomic-level 3D structural studies of macromolecular machines at work inside frozen cells. I also advise anyone pursuing graduate work in the biosciences to receive training in statistics and analysis of Big Data.”
Expanding Access to Global Data
As he stepped into the leadership role of the RCSB PDB, Dr. Burley set out to help broaden the role of the PDB in expanding the frontiers of fundamental biology, biomedicine, energy sciences, and biotechnology. Funded by the National Science Foundation, the National Institutes of Health, and the Department of Energy, the RCSB PDB is the U.S. data center for the global PDB archive of 3D structure data for large biological molecules and supports structural biologists and PDB data users worldwide, giving them open access to PDB data free of charge and without usage restrictions at the RCSB PDB research-focused web portal (RCSB.org). Additionally, educators, students, and the general public can access the RCSB PDB training, outreach, and education-focused web portal (PDB101.RCSB.org) for an exciting exploration of the accumulating knowledge of 3D structure, function, and evolution of the molecules of life.
With just seven protein structures, the PDB was established in 1971 as the first open-access digital data resource in biology and medicine. Over the past fifty-plus years, the archive has grown to more than 200,000 3D structures of proteins and nucleic acids (and their complexes with one another and small-molecule ligands) contributed by structural biologists working on all inhabited continents. “Not only has the total number of PDB structures grown substantially in the past fifty-plus years, but their complexity increases every year,” explains Burley. “The most recent accomplishment of the RCSB PDB is the integration of PDB structures with more than 1 million Computed Structure Models (CSM) coming from the AlphaFold DataBase and the ModelArchive on our RCSB.org research-focused web portal.”
Dr. Burley continues, “Today, RCSB.org delivers both kinds of 3D structure information with clear identification as to data provenance and reliability, allowing us to better serve the 99% of our users who are not experts in structural biology. Extensible and flexible PDB data management and cyberinfrastructure resources developed over the past 50 years (particularly during the last 10 years) made this possible. It was an enormous team effort that depended on the expertise of many talented structural biologists, data scientists, software engineers, and information technology professionals.”
3D biostructure data stored in the PDB have supported notable advances in understanding protein architecture and its role in human and animal health and disease, plants and food and energy production, and global prosperity and sustainability. Historically, structural studies of biomolecules were size limited and focused on isolated proteins. Advances in technology have revealed larger and more complex structures and allowed the examination of a biomolecule’s dynamic behavior to gain a better understanding of function. Using the methods of nuclear magnetic resonance (NMR) spectroscopy, cryo-electron microscopy, and X-ray crystallography, structural biologists can determine the location of each atom relative in a biomolecule.
Data contributed by structural biologists are rigorously validated, expertly biocurated, and then publicly released into the PDB archive, where users can access structures for ribosomes, oncoproteins, drug targets, and even whole viruses. Serving many millions of data consumers worldwide every year, the data stored in the PDB continues to have a profound impact on basic and applied research, discovery of lifesaving drugs, patentable technology discoveries, biotechnology product development, and STEAM education—helping to expand our collective knowledge on a global scale.
PDB Archive
- Managed by International collaboration: US-Asia-Europe
- Contains more than 200,000 structures of proteins, DNA, and RNA
- Used to download > 8 million structure data files per day
- Manages “Big Data” as global Public Good
- Grows at the rate of nearly 10% per year
- The cost to replicate the contents of the PDB archive is estimated at $20 billion
PDB Data
- Used by nearly 500 biological data resources
- Contributed data to >1 million published research papers
- Enables research in subject areas from Agriculture to Zoology
Source: Enabling Breakthroughs in Scientific and Biomedical Research and Education. RCSB PDB.
Building Collaborative Partnerships
With collaboration at the core of the PDB, tapping into the knowledge, experience, and capabilities of other innovators in the research and education space is essential. “We collaborate on various activities pertaining to the Rutgers Drug Discovery and Development Ecosystem and making 3D biostructure information broadly available to our community of scholars,” says Burley. “In December 2021, Rutgers IQB, the RCSB PDB, the Office of Advanced Research Computing (OARC), and Rutgers Office for Research collaborated on delivering a Crash Course, entitled “Enabling Protein Structure Prediction with Artificial Intelligence at Rutgers and Beyond.” Hundreds of participants were attracted from around the world to this event, wherein expert speakers provided a solid foundation on the role of AI/ML in structural biology and showcased ongoing research efforts at Rutgers University. Attendees also had the opportunity to gain hands-on experience with open-source AI tools that predict 3D structures of smaller globular proteins with accuracies comparable to low resolution experimental methods.
To help more institutions within the educational research community gain access to a broad range of collaborative multi-institutional resources, Dr. Burley says they also partner with the Ecosystem for Research Networking (ERN). “I chair the ERN Structural Biology Working Group and foster collaborations between the ERN, Rutgers IQB, and the PDB. One of our most important activities is focused on the Rutgers Cryo-Electron Microscopy and Nanoimaging Facility (RCNF).” The RCNF preserves biological specimens in their native state by vitrification (rapid cooling of liquid medium without ice crystal formation). Then using cryo-transmission electron microscopy, cryo-scanning electron microscopy, or cryo-focused ion beam microscopy, the specimens are imaged and the 3D information is reconstructed through computational analysis of these nanoscale images. Through the partnership with the OARC, RCNF is able to maintain a university-wide installation of certain cryo-EM software, affording more users the opportunity to use this cutting-edge technology.
Rutgers has long partnered with Edge for network services and delivery of superior network connectivity experience to their users. Recently, Rutgers further upgraded this experience by migrating one of their enterprise Internet connections to the Edge optical fiber network, EdgeNet, at 100 Gbps. “In calendar year 2022, RCSB.org was accessed by more than 7 million unique IP addresses, coming from virtually every sovereign country recognized by the United Nations,” says Burley. “PDB data are also used by millions more researchers, educators, and students who access the information from nearly 500 trusted external data resources that reuse and repackage PDB data. Additionally, every major biopharmaceutical company maintains its own copy of the PDB archive inside the organizational firewall. The PDB and Rutgers IQB benefit greatly from high performance connectivity provided through the Regional Research and Education network at Edge.”
“The incredible progress of our academic partners across the research community never ceases to amaze me. Fast and reliable access to data and technology is fundamental for research and development. It is exciting for Edge to be able to help the region’s most creative minds discover breakthroughs and solutions to some of the world’s greatest scientific challenges,” mentions Dr. Forough Ghahramani, Assistant Vice President for Research, Innovation, and Sponsored Programs at Edge.
Insulin Release by David S. Goodsell, RCSB Protein Data Bank and Scripps Research.
doi: 10.2210/rcsb_pdb/goodsell-gallery-044
This painting depicts one of the few examples of a protein crystal with a biological function. Insulin is stored in pancreatic beta cells in the form of small crystals, carried inside specialized vesicles. When insulin is needed after meals, these vesicles fuse with the cell membrane and the crystals dissolve, releasing insulin into the bloodstream. The painting shows the vesicle in the process of releasing insulin. The insulin crystal is at the top in yellow, the fused vesicle membrane and cell membrane are in green, and the extracellular matrix that surrounds the beta cell is at the bottom in tans and browns.
Revealing New Opportunities
With today’s scientific landscape looking much different than it was two or three decades ago, Dr. Burley is looking forward to the new opportunities that lay ahead. “My IQB and PDB colleagues and I are currently focused on four areas in the next ten years. First is supporting the continuous growth in the number and complexity of 3D biostructures stored in the PDB archive, which has been doubling in size every 6-8 years. We must preserve and distribute this information as broadly as possible at no charge and with no limitations on usage. Second, we are seeing explosive growth in the number of publicly available CSMs (approaching 1 billion, with the recent release of ~600 million predicted structures of proteins by Meta AI). We must make more of this information accessible alongside PDB structures to maximize its value for the research and education communities.”
Dr. Burley continues, “With the rapid growth in the number and diversity of RCSB.org users, coming from across fundamental biology, biomedicine, energy sciences, bioengineering, computer science, statistics, chemistry, physics, and mathematics, we must also understand and continue to augment the utility of the RCSB.org web portal to meet the needs of these communities. Finally, we must ensure that the Rutgers IQB and PDB (via RCSB.org and PDB101.RCSB.org) have the broadest possible societal impact by creating opportunities to engage and foster the careers of under-represented minority students, educators, and researchers. I view all challenges as opportunities. Experimental structural biology has a very bright future; helping to enable further breakthroughs in basic and applied research and education.”